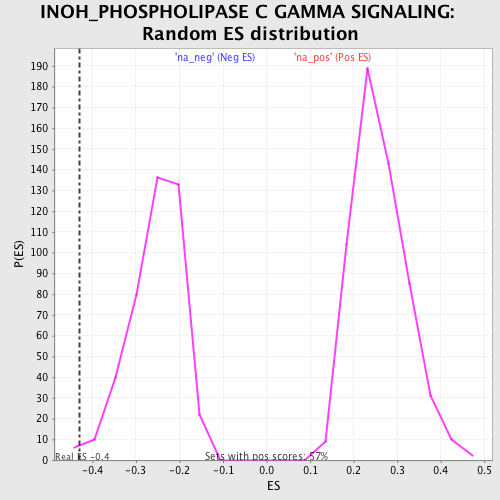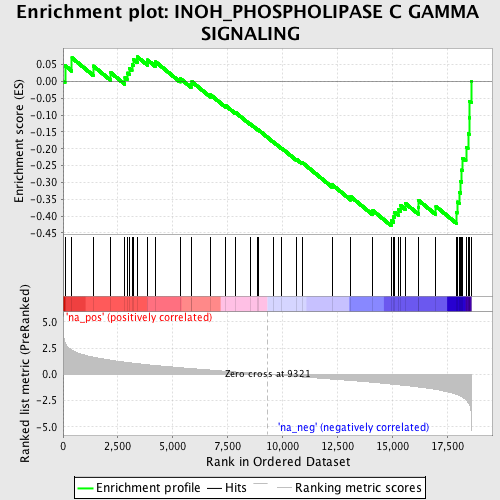
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_PHOSPHOLIPASE C GAMMA SIGNALING |
| Enrichment Score (ES) | -0.42927 |
| Normalized Enrichment Score (NES) | -1.6833725 |
| Nominal p-value | 0.007025761 |
| FDR q-value | 0.63271105 |
| FWER p-Value | 0.98 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TYK2 | 88 | 3.032 | 0.0472 | No | ||
| 2 | JAK3 | 404 | 2.299 | 0.0696 | No | ||
| 3 | HRAS | 1373 | 1.641 | 0.0456 | No | ||
| 4 | ITPR3 | 2174 | 1.342 | 0.0255 | No | ||
| 5 | PRKCQ | 2817 | 1.156 | 0.0108 | No | ||
| 6 | RELA | 2917 | 1.129 | 0.0248 | No | ||
| 7 | NFKBIE | 3027 | 1.102 | 0.0378 | No | ||
| 8 | ZAP70 | 3141 | 1.073 | 0.0501 | No | ||
| 9 | CSK | 3210 | 1.058 | 0.0645 | No | ||
| 10 | MAPK3 | 3384 | 1.017 | 0.0727 | No | ||
| 11 | ARAF | 3845 | 0.911 | 0.0635 | No | ||
| 12 | NFKBIB | 4219 | 0.840 | 0.0578 | No | ||
| 13 | PRKCA | 5331 | 0.637 | 0.0089 | No | ||
| 14 | LCK | 5834 | 0.552 | -0.0087 | No | ||
| 15 | PRKCI | 5870 | 0.546 | -0.0012 | No | ||
| 16 | SRC | 6721 | 0.404 | -0.0401 | No | ||
| 17 | FOSB | 7400 | 0.293 | -0.0716 | No | ||
| 18 | PRKCH | 7861 | 0.227 | -0.0924 | No | ||
| 19 | FOSL1 | 8533 | 0.121 | -0.1265 | No | ||
| 20 | TEC | 8871 | 0.065 | -0.1435 | No | ||
| 21 | NFKB1 | 8893 | 0.063 | -0.1436 | No | ||
| 22 | MAP2K1 | 9587 | -0.041 | -0.1802 | No | ||
| 23 | PRKCD | 9961 | -0.109 | -0.1984 | No | ||
| 24 | FOS | 10648 | -0.208 | -0.2318 | No | ||
| 25 | JAK1 | 10897 | -0.252 | -0.2408 | No | ||
| 26 | PTK2B | 12267 | -0.449 | -0.3069 | No | ||
| 27 | MAPK9 | 13114 | -0.579 | -0.3425 | No | ||
| 28 | KRAS | 14116 | -0.757 | -0.3834 | No | ||
| 29 | MAP2K4 | 14968 | -0.928 | -0.4134 | Yes | ||
| 30 | SYK | 15062 | -0.946 | -0.4022 | Yes | ||
| 31 | NRAS | 15105 | -0.954 | -0.3881 | Yes | ||
| 32 | JUN | 15285 | -0.989 | -0.3808 | Yes | ||
| 33 | MAP2K2 | 15389 | -1.012 | -0.3690 | Yes | ||
| 34 | JAK2 | 15596 | -1.055 | -0.3620 | Yes | ||
| 35 | RASGRP1 | 16193 | -1.200 | -0.3735 | Yes | ||
| 36 | PRKCE | 16205 | -1.204 | -0.3535 | Yes | ||
| 37 | NFKBIA | 16992 | -1.433 | -0.3713 | Yes | ||
| 38 | ITK | 17918 | -1.886 | -0.3888 | Yes | ||
| 39 | MAPK10 | 17957 | -1.913 | -0.3580 | Yes | ||
| 40 | FOSL2 | 18055 | -2.012 | -0.3288 | Yes | ||
| 41 | BTK | 18115 | -2.064 | -0.2966 | Yes | ||
| 42 | ITPR1 | 18169 | -2.124 | -0.2630 | Yes | ||
| 43 | FYN | 18200 | -2.163 | -0.2276 | Yes | ||
| 44 | PRKCZ | 18363 | -2.401 | -0.1952 | Yes | ||
| 45 | MAPK8 | 18456 | -2.662 | -0.1545 | Yes | ||
| 46 | RAF1 | 18508 | -2.852 | -0.1084 | Yes | ||
| 47 | PLCG1 | 18519 | -2.887 | -0.0595 | Yes | ||
| 48 | PTK2 | 18604 | -3.772 | 0.0006 | Yes |